An Integrated Molecular, Cellular
and Genomic Investigation of DNA Double-Strand Break Repair in Bacteria | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Our laboratory aims to
dissect the molecular mechanism of DNA double-strand break repair (DSBR) as it
occurs in living E. coli cells. Despite the decades of research on bacterial
DSBR, the application of new technologies to sophisticated in vivo systems for generating DNA breaks is generating new and
unexpected insight into this fundamental macromolecular reaction.
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
DNA repair pathways are remarkably well conserved from bacteria, through yeasts to humans (see Table below for a list of equivalent recombination proteins). |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Table: Recombination proteins
in E. coli, S. cerevisiae and Human
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
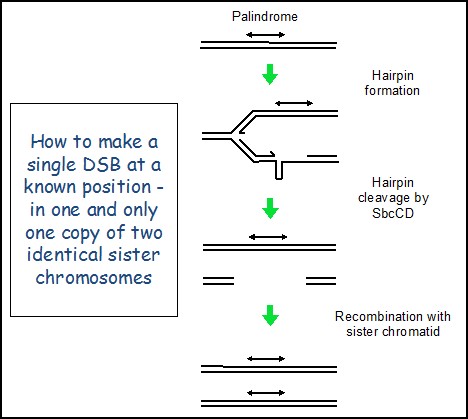
Creating a repairable DNA double strand break in E. coli The primary role of genetic recombination in DSBR is the repair of post-replicative breaks using the identical genetic information present on an undamaged sister chromosome. However, this has been problematic to investigate at the molecular level in any system because the two recombining partners are genetically identical. This means that it is difficult to target a DNA double-strand break (DSB) to one sister chromosome at a specific location without risking cleavage of the other sister chromosome at the same site, thereby removing the intact template for repair. We have overcome this problem by developing a system in E. coli where the cleavage by SbcCD nuclease of a DNA hairpin formed on the lagging-strand template at the site of a chromosomal palindrome (lacZ::pal) initiates DSBR following DNA replication, as shown here. Remarkably, the DSBR event is so efficient that doubling time of these cells is close to unaffected and they only suffer a 0.06% loss of fitness per generation, despite inducing the SOS checkpoint response and increasing in size.
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Visualisation of E. coli cells subjected or not to DSB video no DSB | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
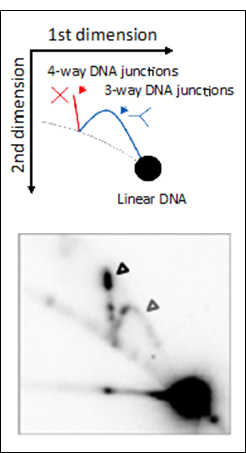
Molecular investigation of DSBR in | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Cellular and genomic investigation of DSBR in E. coli Our system of live cell imaging is enabling us to visualise the two sides of the DSB (as LacI-Cerulean and TetR-YPet foci) and their association with key proteins as they undergo DSBR. In addition, we have developed genomic technologies (ChIP-seq and genomic sequencing) to investigate of chromosomal DSBR.
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||